BioNMR & Biophysical Characterization
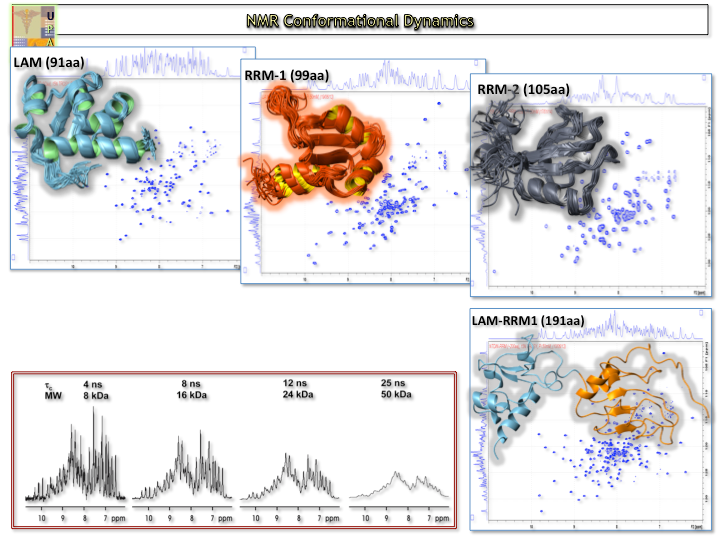
Protein Conformational Dynamics – Structural Biochemistry
The group exhibit expertise and collaborate with many other national or EU research group (in academia or in private sectors) in Biomolecular NMR methods and application to :
-
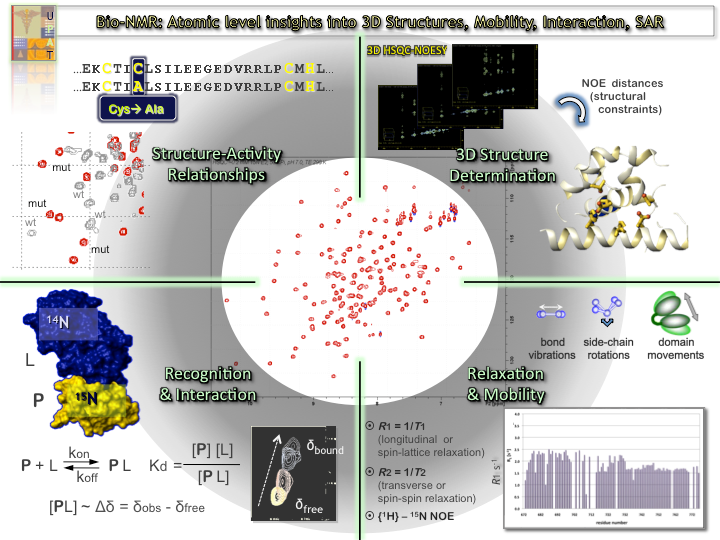
High-resolution structure determination of protein drug-targets with molecular weight above 10 kDa, and atomic-level insight in protein and protein-protein/RNA/DNA/drug complexes dynamics, interaction and determination of binding affinity
-
Protein dynamics using {1H}–15N heteronuclear NOEs, 15N longitudinal (R1) and transverse (R2) relaxation rates on 15N-labelled protein samples and chemical exchange saturation transfer (CEST)
-
Coupled fast acquisition and automated assignment techniques with unsupervised structure determination of proteins that reduces the time of the entire proceedure from months to weeks.
-
Time-resolved kinetics and fast acquisition of 2D fingerprint, diagnostic, 1H-15N HSQC spectra (~1 sec), eventually crucial for monitoring protein-protein/peptide/small-molecules transient complexes, folding/unfolding processes and mechanistic studies, protein-drug screening applicable in drug-design process.
-
Use of reduced protein amounts and investigation of low concentration protein samples (since numerous proteins exhibit low expression yields, while others tend to homo- or hetero-oligomerize or aggregate in medium to high concentrations, i.e. >0.4-0.6 mM).
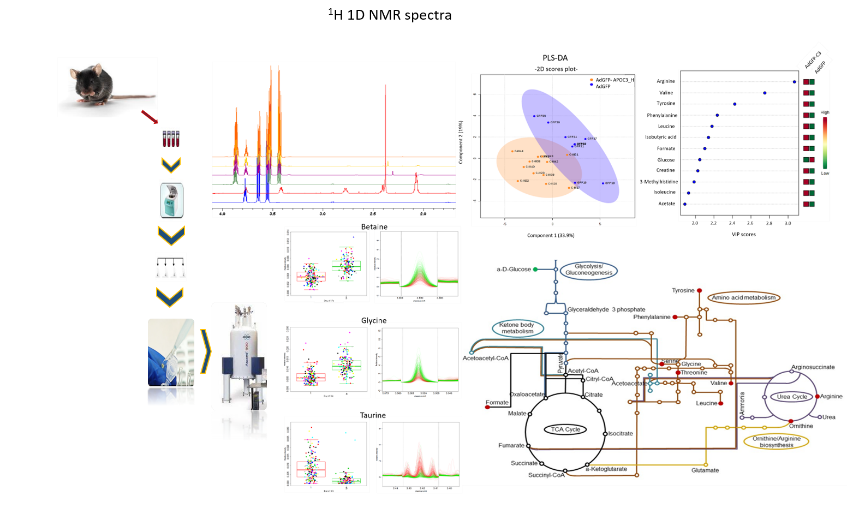
NMR Metabolomics
Analysis of biological fluids include metabolite recording and quantification in a single experimental run and evaluation of metabolite level alterations.
Methodology applied:
-
(a) Targeted metabolic identification profiling experiments (set of 1D & 2D NMR spectra/sample), following the concentration levels of a limited set of metabolites. Specific biochemical pathways are highlighted and integration of other ‘’-omic’’ approaches can be applied.
-
(b) Untargeted metabolite identification and metabolic fingerprinting (1H 1D NMR spectrum per sample), aiming a comprehensive and simultaneous analysis of a wide variety of compounds.
Biophysical Characterization
Our lab produce high-quality protein samples in high-yield and in many cases in labeled form (2H, 13C, 15N). A wide range of analytical tools are used in order to assess the quality of the produced recombinant proteins elucidate their biophysical properties, such as UV-VIS spectroscopy, Fluorescence Spectroscopy, Circular Dichroism (CD) and Size Exclusion Chromatography – Multiple Angle Lights Scattering (SEC-MALS) to evaluate and characterize in depth their structure, function and stability.
-
UV-VIS spectroscopy: It provides the opportunity to determine the concentration of Protein, DNA or RNA in different samples. Furthermore, it assists us to identify the oxidation state of several elements.
-
Fluorescence Spectroscopy is an attractive tool, which can provide information at a nanoscopic level, with exceptional sensitivity, for analyzing and acquiring data regarding to the structure and properties of biomolecules, (i.e. protein interaction) with high precision.
-
Circular Dichroism: The content of protein secondary structure can be determined by CD spectroscopy in the far-UV spectral region (190-260 nm). At these wavelengths the chromophore is the peptide bond, and the signal arises when it is in a regular, folded environment. Alpha-helical, beta-sheet, and random coil structures give rise to a characteristic shape and magnitude of CD spectrum. The approximate fraction of each secondary structure type presents in a protein solution can thus be determined by analyzing its far-UV CD spectrum.
-
SEC Multiple Angle Lights Scattering: SEC MALS can be used to analyze protein samples far beyond the study of their aggregation state and their sample stability. This technique is capable to provide valuable biophysical data, such as the hydrodynamic radius of the molecule, to determine the absolute molar mass (from hundreds of Daltons to million Daltons) and characterize the shape and the size (10nm to 300nm) of the vesicles, to estimate the protein concentration in solutions and characterize protein/peptide conjugates. As an absolute technique for determining molar mass and size in solution, SEC-MALS offers an advanced characterization overcoming the many limitations of column calibration.
Thermodynamic & Kinetics
Moreover, our lab applies thermodynamic techniques such as Isothermal Titration Calorimetry (ITC) with the aim of studying biomolecular interactions (between proteins, nucleic acids and small molecules) and ligand binding. This method provides accurately the affinity constant (Kd), the stoichiometry (n), the enthalpy (ΔH) and the entropy of biomolecular interactions in a single experiment.
This technique:
-
increases the understanding of the molecular recognition process
-
identifies and optimizes potential leads on their binding characteristics